The attached Summary Protocol and Full Protocol have specifically been written by Dr Nat Milton of Leeds Trinity University plus Neurodelta Ltd, who has used antisense peptide techniques for both academic research and commercial activities. The methods detailed in the protocol are aimed at University students plus academic staff for use in non-commercial activities. The main method is based around antisense peptide screening [1] and has been adapted to protein sequence database screening [2]. Support for these Bioinformatics protocols from Dr Milton is available directly via Email.
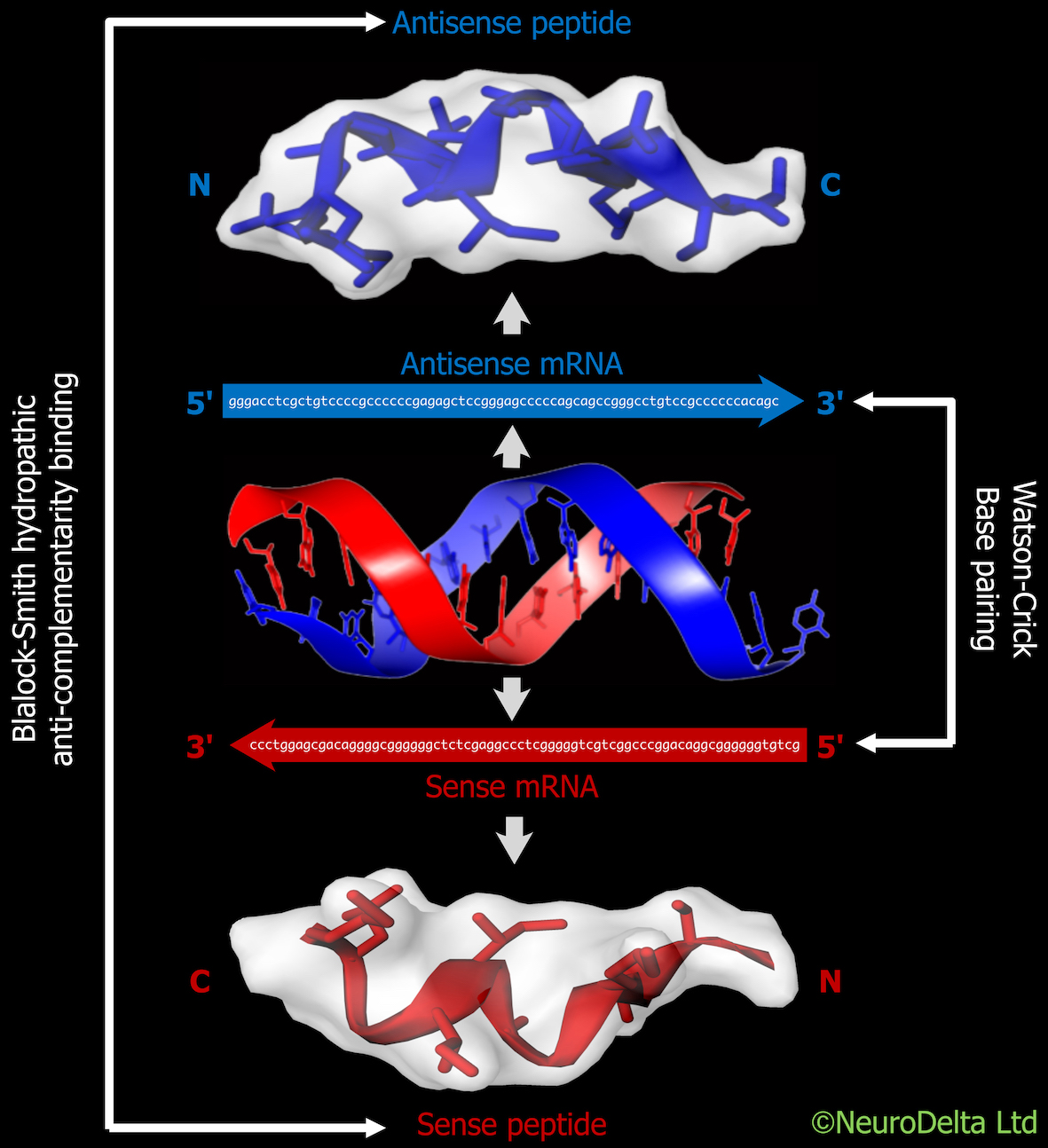
The protocol details how to use an online method (using a Python script or Word version which can be used in an online Python Compiler or a downloaded version of Python 3) to generate antisense peptide sequences from a target protein mRNA sequence and has been based on the methods outlined in [2],[3],[4]. Methods using the antisense peptides in protein BLAST searches to identify potential protein interactions with the target protein are included. Protein-protein docking of 3D models can be used to predict the structure of complexes between the proteins identified from the antisense peptide screening [5],[6].
Bioinformatics Protocols Site Support:
Email: info@bioinformatics-protocols.com